
| SOFTWARE |
BioRedox-MT3D v1 is a 3-D reactive transport model which was developed by Grant Carey of Porewater Solutions, with support from Dr. Paul Van Geel (Carleton University) and J. Richard Murphy (Conestoga-Rovers & Associates or CRA), and was developed with partial funding provided by CRA.
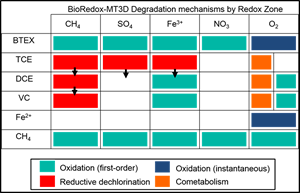
BioRedox-MT3D provides a flexible reaction framework which allows for optional simulation of coupled oxidation-reduction reactions (e.g. electron donors and electron acceptors), sequential reductive dehalogenation or radionuclide decay, cometabolic degradation, and substrate-dependent or inhibitor-dependent reaction rates.
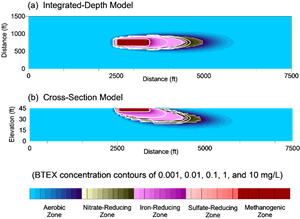
BioRedox-MT3D also uses a unique visualization system which facilitates plotting redox zones and contaminant of concern concentrations on a single figure.
BioRedox-MT3D v1 is a public domain model and may be freely copied or re-distributed. Source code, User manuals, and example simulations are available below for download.